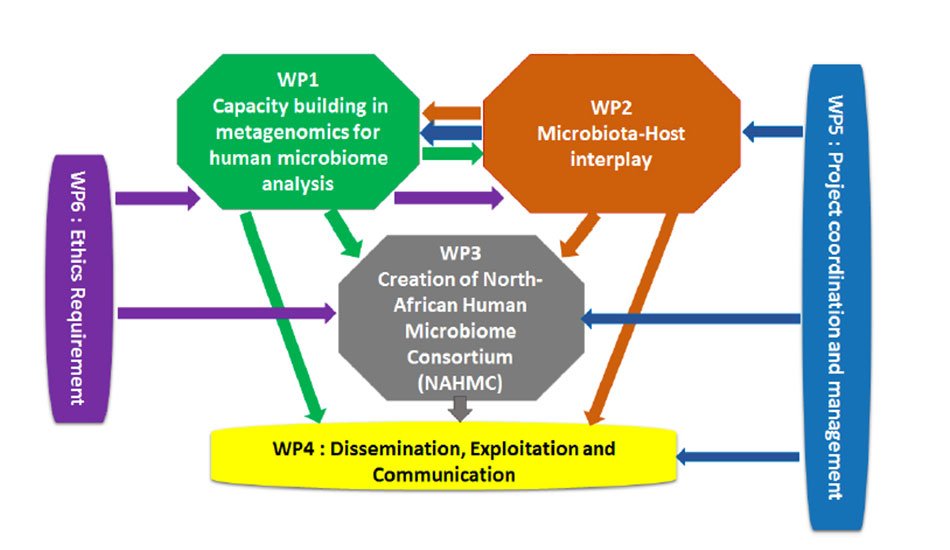
WP1: Capacity Building in metagenomics for human microbiome analysis
-
Task 1.1. Adoption the state of the art standardization protocols
We aim to adopt standardized protocols for collecting stool samples from Tunisian healthy and CRC patients. It is important to optimize the fecal sampling pipeline in a prudent way to get high quality metagenomic DNA for an unbiased NGS and bioinformatic analysis. The fecal collection kit, transportation condition, storage status, and DNA extraction method have to be done according to standardized protocols. -
Task 1.2. Strengthening skills in metagenomics approaches
This task aims to provide a detailed understanding of cutting-edge techniques used in microbiome analysis with an emphasis on the analysis of biological samples. Microbial DNA extracted from stool samples using standardized protocol will be analysed by 16S rRNA and rpoB gene sequencing, then raw data will be processed using bioinformatics tools. -
Task 1.3. Dysbiosis of gut microbiota and colorectal cancer
Colorectal Cancer (CRC) is one of the most common malignancies worldwide and its pathogenesis is a complex and multifactorial process with accumulation of various genetic and epigenetic alterations. It was demonstrated that quantitative and qualitative changes in the intestinal microbiome, contribute to the development of CRC. Therefore, monitoring changes in the gut microbiome during CRC development could be an easy diagnostic tool, a useful biomarker for adapting the therapy in CRC patients. In this subtask, a pilot study including 10 Tunisian CRC patients to investigate by 16S RNA amplicon sequencing the gut microbiome. Data will be analyzed by bioinformatics tools and bacterial communities between cancer patients and healthy individuals will be compared to identify bacterial species present in the gut of cancer patients. -
Task 1.4. Audit to validate the implemented procedures for microbiome analysis
We will submit ourselves to a standard audit by one of the global initiatives on microbiome to check our alignment to international procedures. This will enable us to raise our profile as a credible partner able to integrate international consortium, to share data and to take part in common efforts on the human gut microbiome for the development of personalized treatment in the future.
WP2: Microbiota-Host interplay
-
Task 2.1. Capacity building in Functional Metagenomics
To further characterize the microbiota-host interplay the strategies will consist to i) construction of metagenomics libraries and their screening, ii) analysis of commensal bacteria effects and iii) study of the metagenomics catalogs aiming the identification of target genes. To explore the effects of bacteria/genes, different high throughput in vitro systems including, cell reporter systems and protein activity measurements and also animal models (mice, rabbit and pigs) will be used. -
Task 2.2 Improvement skills in Metabolomics
Metabolomics is a powerful technique that detects hundreds of small molecules present in biological system such as fecal. Thus, examining the fecal metabolomes serve as a strategy for understanding the interactions between diet, human metabolism, and the gut microbiota composition in health and disease. In this regard, to analyse metabolites in biological samples, several mass spectrometry-based techniques (MS) and nuclear magnetic resonances spectroscopy (NMR) and GC-MS have been employed. Consequently, characterization of metabolic phenotypes can enable personalized medicine -
Task 2.3. Development of a teaching module on human microbiome
A teaching module on human microbiome and its relationship with health/diseases will be developed and dedicated to students at Master level (USFAX and NA universities). This module aims to provide a detailed understanding of cutting-edge techniques used in microbiome analysis with an emphasis on functional metagenomics and clinical applications. This course will be available on the Moodle plateform of the Faculty of Medecine of Sfax.
WP3: Creation of North-African Human Microbiome Consortium (NAHMC)
-
Task 3.1. Preparation of an agreement for the creation of the North-African Human Microbiome Consortium
An agreement summarizing criteria of the NAHMC (data release, quality assessment, standardization of procedures and protocols, consent of participants) will be drafted and approved by the NA universities members. Upon creation, a 5-year strategy will be discussed and an action plan outlining the objectives of the NAHMC will be prepared. -
Task 3.2. Coordination and planning the NAHMC strategy and action plan
Thanks to the expertise in the field of microbiome, that USFAX will acquire with the support of EU partners, USFAX will occupy a leading position in NA and will coordinate the activities of this consortium. Among its activities, the NAHMC will organize meetings, integrate the International Human Microbiome Consortium (IHMC), participate to international meetings. Networking activities will be also organized to spread good practices and to encourage complementarity between the members of the consortium by widening participation of ER and ESR from Tunisian and NA universities with respect of gender balance.
WP4: Dissemination, Exploitation and Communication
- Website creation
- Peer-reviewed Publication on human microbiome in high impact-factor journals
- Integration into EU and international consortiums on human microbiome
- Organisation of academic conferences on human microbiome
- LinkedIn” will be also used to establish networks on microbiome analysis.
- Publication of a periodic newsletter “MICAfrica” at the website describing all activities and the event that will be led by MICAfrica
- Dissemination through different networks and media centers such as “Cordis Wire”
- Use of social media (Facebook, Twitter, Newsletter)
- Edition of quarterly scientific bulletins (“MICAfrica”)
- Biannual Report
- Printed communications
- Awarness compaign (Public talks , direct personal contacts)
- Creation of Microbiome database
- Creation and coordination of NACHM
- Integration into EU and international consortiums on human microbiome
- On line module on the human microbiome (MHM)
- Master’s degree on HM as part of the Erasmus + KA2 program
WP5: Project Coordination and Management
During the Kick Off meeting (KoM) the management structure and the Steering Committee (SC) is established. The SC is composed by the Contact Persons of each member of the consortium and administrative/technical staff (PC, Project manager and WPL) who are actively involved in all activities.
An advisor board is associated to the management structure. It comprise a high level international experts that provide advices and quality control of procedure standardization. This advisor board interact tightly with SC and PC.
Transparency and accountability are core principles in the decision-making process and project management. SC and PC make their best attempts to resolve conflicts and to take appropriate decisions at the right time. Each conflict will be resolved effectively with a very open and collaborative mind. The SC will focus on resolving the conflict and finding the best alternative for the team and the creative solution acceptable to everyone. This collaborative decision-making strategy leads to a win-win outcome.

WP6: Ethics requirements
-
D6.1 : H
Regarding the collection of samples from humans: The procedures and criteria that will be used to identify/ recruit research participants must be provided; The informed consent procedures that will be implemented for the participation of humans must be provided; Templates of the informed consent/assent forms and information sheets (in language and terms intelligible to the participants) must be kept on file; Details on incidental findings policy must be provided. -
D6.2 : POPD
The beneficiary must check if special derogations pertaining to the rights of data subjects or the processing of genetic, biometric and/or health data have been established under the national legislation of the country where the research takes place and submit a declaration of compliance with respective national legal framework(s). The host institution must confirm that it has appointed a Data Protection Officer (DPO) and the contact details of the DPO are made available to all data subjects involved in the research. For host institutions not required to appoint a DPO under the GDPR, it must be confirmed that a detailed data protection policy for the project is available and kept on file. The beneficiary must explain how all of the data they intend to process is relevant and limited to the purposes of the research project (in accordance with the ‘data minimisation ‘principle).A description of the technical and organisational measures that will be implemented to safeguard the rights and freedoms of the data subjects/ research participants must be provided. In case personal data are transferred from the EU to a non-EU country or international or-ganisation, confirmation that such transfers are in accordance with Chapter V of the General Data Protection Regulation 2016/679, must be provided. In case personal data are transferred from a non-EU country to the EU (or another third state), confirmation that such transfers comply with the laws of the country in which the data was collected must be provided. In case of further processing of previously collected personal data, an explicit confirmation that the beneficiary has lawful basis for the data processing and that the appropriate technical and organisational measures are in place to safeguard the rights of the data subjects must be provided.

